Training: Quantum case¶
In this tutorial we will train a lambeq model to solve the relative pronoun classification task presented in [LPM+23]. The goal is to predict whether a noun phrase contains a subject-based or an object-based relative clause. The entries of this dataset are extracted from the RelPron dataset [RMPC16].
We will use an IQPAnsatz to convert string diagrams into quantum circuits. The pipeline uses tket as a backend.
If you have already gone through the classical training tutorial, you will see that there are only minor differences for the quantum case.
Preparation¶
We start with importing NumPy and specifying some training hyperparameters.
import os
import warnings
warnings.filterwarnings('ignore')
os.environ['TOKENIZERS_PARALLELISM'] = 'true'
Note
We disable warnings to filter out issues with the tqdm package used in jupyter notebooks. Furthermore, we have to specify whether we want to use parallel computation for the tokenizer used by the BobcatParser. None of the above impairs the performance of the code.
import numpy as np
BATCH_SIZE = 10
EPOCHS = 100
SEED = 2
Input data¶
Let’s read the data and print some example sentences.
def read_data(filename):
labels, sentences = [], []
with open(filename) as f:
for line in f:
t = int(line[0])
labels.append([t, 1-t])
sentences.append(line[1:].strip())
return labels, sentences
train_labels, train_data = read_data('../examples/datasets/rp_train_data.txt')
val_labels, val_data = read_data('../examples/datasets/rp_test_data.txt')
train_data[:5]
['organization that church establish .',
'organization that team join .',
'organization that company sell .',
'organization that soldier serve .',
'organization that sailor join .']
Targets are represented as 2-dimensional arrays:
train_labels[:5]
[[1, 0], [1, 0], [1, 0], [1, 0], [1, 0]]
Creating and parameterising diagrams¶
The first step is to convert sentences into string diagrams.
Note
We know that the specific dataset only consists of noun phrases, hence, we reduce potential parsing errors by restricting the parser to only return parse trees with the root categories N (noun) and NP (noun phrase).
from lambeq import BobcatParser
parser = BobcatParser(root_cats=('NP', 'N'), verbose='text')
raw_train_diagrams = parser.sentences2diagrams(train_data, suppress_exceptions=True)
raw_val_diagrams = parser.sentences2diagrams(val_data, suppress_exceptions=True)
Tagging sentences.
Parsing tagged sentences.
Turning parse trees to diagrams.
Tagging sentences.
Parsing tagged sentences.
Turning parse trees to diagrams.
Filter and simplify diagrams¶
We simplify the diagrams by calling normal_form() and filter out any diagrams that could not be parsed.
train_diagrams = [
diagram.normal_form()
for diagram in raw_train_diagrams if diagram is not None
]
val_diagrams = [
diagram.normal_form()
for diagram in raw_val_diagrams if diagram is not None
]
train_labels = [
label for (diagram, label)
in zip(raw_train_diagrams, train_labels)
if diagram is not None]
val_labels = [
label for (diagram, label)
in zip(raw_val_diagrams, val_labels)
if diagram is not None
]
Let’s see the form of the diagram for a relative clause on the subject of a sentence:
train_diagrams[0].draw(figsize=(9, 5), fontsize=12)
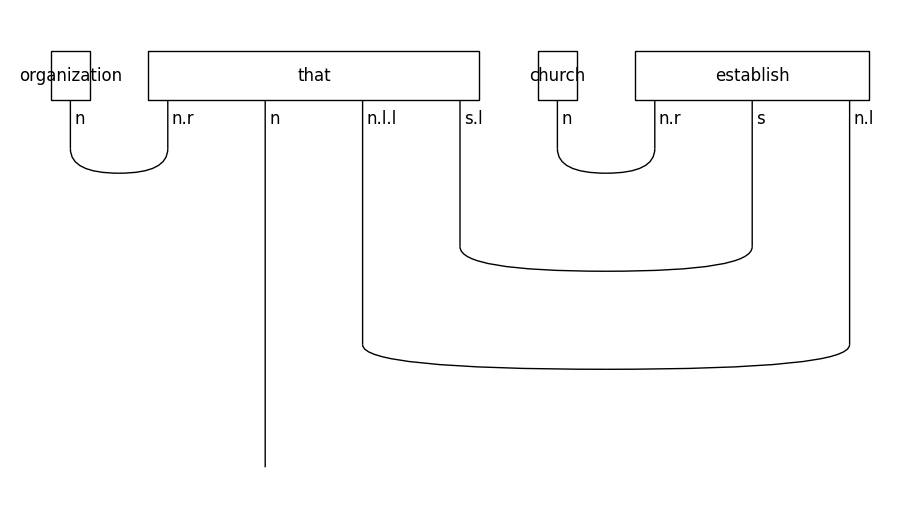
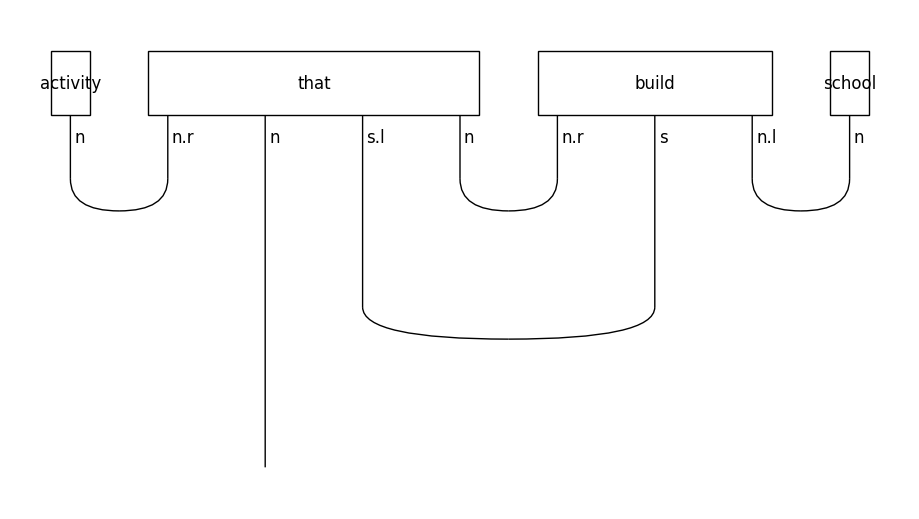
In object-based relative clauses the noun that follows the relative pronoun is the object of the sentence:
train_diagrams[-1].draw(figsize=(9, 5), fontsize=12)
Create circuits¶
In order to run the experiments on a quantum computer, we need to apply to string diagrams a quantum ansatz. For this experiment, we will use an IQPAnsatz, where noun wires (n) are represented by a one-qubit system, and sentence wires (s) are discarded (since we deal with noun phrases).
from lambeq import AtomicType, IQPAnsatz, RemoveCupsRewriter
ansatz = IQPAnsatz({AtomicType.NOUN: 1, AtomicType.SENTENCE: 0},
n_layers=1, n_single_qubit_params=3)
remove_cups = RemoveCupsRewriter()
train_circuits = [ansatz(remove_cups(diagram)) for diagram in train_diagrams]
val_circuits = [ansatz(remove_cups(diagram)) for diagram in val_diagrams]
train_circuits[0].draw(figsize=(9, 10))
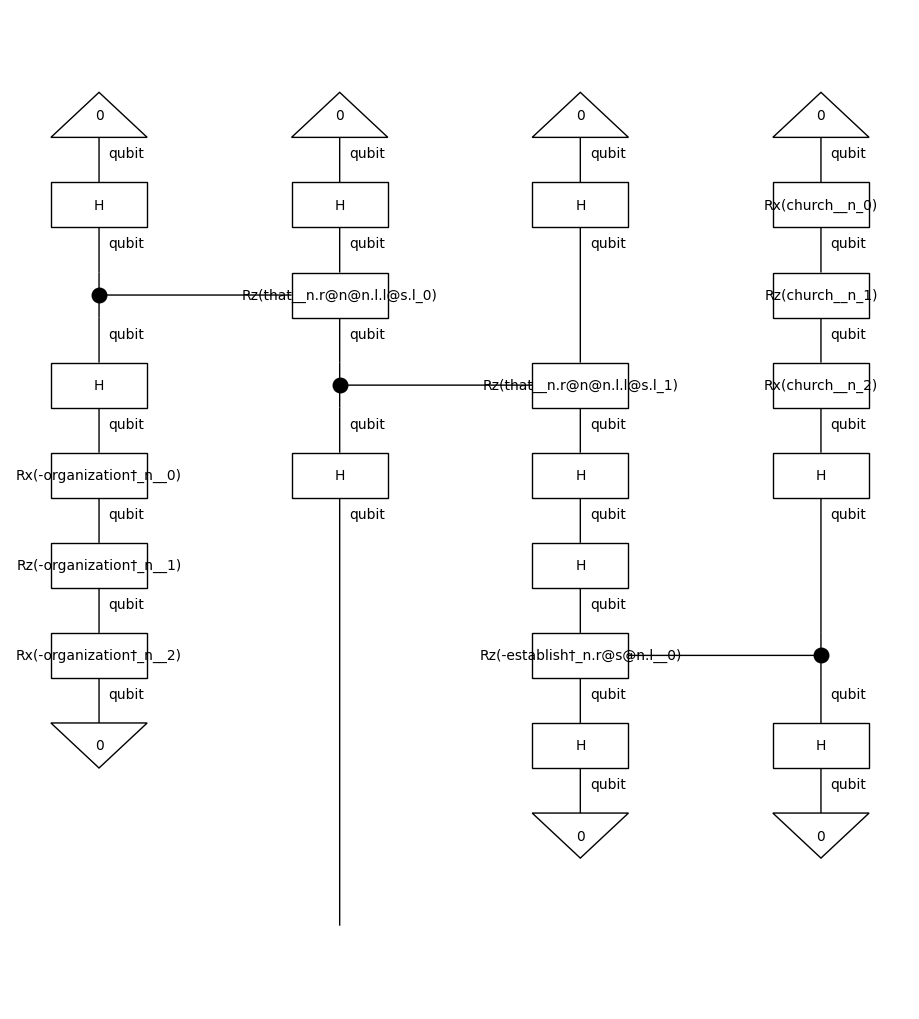
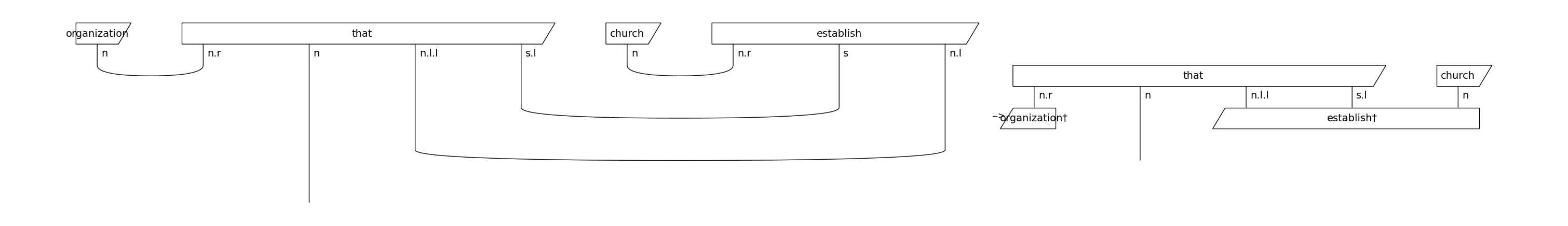
Note that we remove the cups before parameterising the diagrams. By doing so, we reduce the number of post-selections, which makes the model computationally more efficient. The effect of cups removal on a string diagram is demonstrated below:
from lambeq.backend import draw_equation
original_diagram = train_diagrams[0]
removed_cups_diagram = remove_cups(original_diagram)
draw_equation(original_diagram, removed_cups_diagram, symbol='-->', figsize=(30, 6), asymmetry=0.3, fontsize=14)
Training¶
Instantiate the model¶
We will use a TketModel, which we initialise by passing all diagrams to the class method
TketModel.from_diagrams(). The TketModel needs a backend configuration dictionary passed as a keyword argument to the initialisation method. This dictionary must contain entries for backend, compilation and shots. The backend is provided by pytket-extensions. In this example, we use Qiskit’s AerBackend with 8192 shots.
from pytket.extensions.qiskit import AerBackend
from lambeq import TketModel
all_circuits = train_circuits + val_circuits
backend = AerBackend()
backend_config = {
'backend': backend,
'compilation': backend.default_compilation_pass(2),
'shots': 8192
}
model = TketModel.from_diagrams(all_circuits, backend_config=backend_config)
Note
The model can also be instantiated by calling TketModel.from_checkpoint(), in case a pre-trained checkpoint is available.
Define loss and evaluation metric¶
We use standard binary cross-entropy as the loss. Optionally, we can provide a dictionary of callable evaluation metrics with the signature metric(y_hat, y).
from lambeq import BinaryCrossEntropyLoss
# Using the builtin binary cross-entropy error from lambeq
bce = BinaryCrossEntropyLoss()
acc = lambda y_hat, y: np.sum(np.round(y_hat) == y) / len(y) / 2 # half due to double-counting
eval_metrics = {"acc": acc}
Initialise trainer¶
In lambeq, quantum pipelines are based on the QuantumTrainer class. Furthermore, we will use the standard lambeq SPSA optimizer, implemented in the SPSAOptimizer class. This needs three hyperameters:
a: The initial learning rate (decays over time),c: The initial parameter shift scaling factor (decays over time),A: A stability constant, best choice is approx. 0.01 * number of training steps.
from lambeq import QuantumTrainer, SPSAOptimizer
trainer = QuantumTrainer(
model,
loss_function=bce,
epochs=EPOCHS,
optimizer=SPSAOptimizer,
optim_hyperparams={'a': 0.05, 'c': 0.06, 'A':0.001*EPOCHS},
evaluate_functions=eval_metrics,
evaluate_on_train=True,
verbose='text',
log_dir='RelPron/logs',
seed=0
)
Create datasets¶
To facilitate data shuffling and batching, lambeq provides a native Dataset class. Shuffling is enabled by default, and if not specified, the batch size is set to the length of the dataset.
from lambeq import Dataset
train_dataset = Dataset(
train_circuits,
train_labels,
batch_size=BATCH_SIZE)
val_dataset = Dataset(val_circuits, val_labels, shuffle=False)
Train¶
We can now pass the datasets to the fit() method of the trainer to start the training. Here, we perform early stopping if the validation accuracy doesn’t improve within the specified early_stopping_interval epochs.
trainer.fit(train_dataset, val_dataset,
early_stopping_criterion='acc',
early_stopping_interval=5,
minimize_criterion=False)
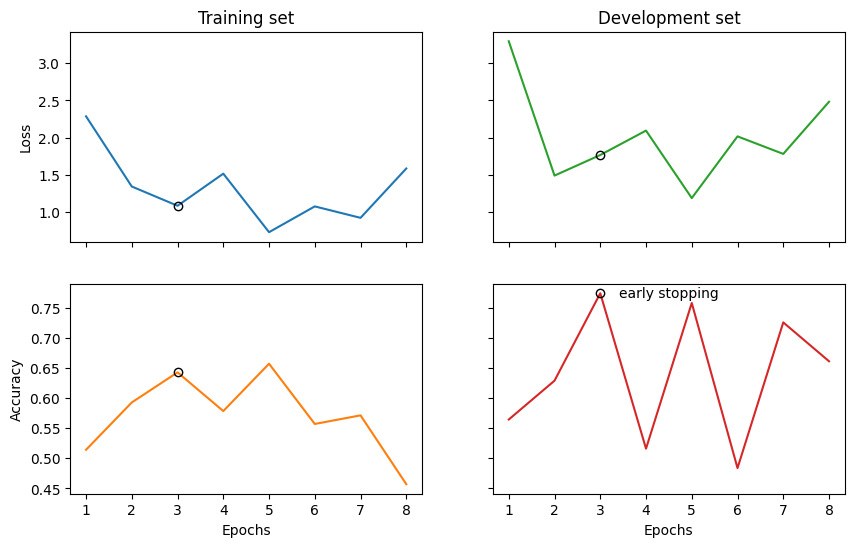
Epoch 1: train/loss: 0.9327 valid/loss: 3.2976 train/time: 13.22s valid/time: 2.94s train/acc: 0.5143 valid/acc: 0.5645
Epoch 2: train/loss: 2.1623 valid/loss: 1.4883 train/time: 14.20s valid/time: 2.83s train/acc: 0.5929 valid/acc: 0.6290
Epoch 3: train/loss: 0.2750 valid/loss: 1.7657 train/time: 13.35s valid/time: 3.04s train/acc: 0.6429 valid/acc: 0.7742
Epoch 4: train/loss: 1.0235 valid/loss: 2.0944 train/time: 12.91s valid/time: 2.75s train/acc: 0.5786 valid/acc: 0.5161
Epoch 5: train/loss: 0.7471 valid/loss: 1.1852 train/time: 13.02s valid/time: 2.87s train/acc: 0.6571 valid/acc: 0.7581
Epoch 6: train/loss: 0.9657 valid/loss: 2.0165 train/time: 12.81s valid/time: 2.91s train/acc: 0.5571 valid/acc: 0.4839
Epoch 7: train/loss: 0.8952 valid/loss: 1.7803 train/time: 12.73s valid/time: 3.00s train/acc: 0.5714 valid/acc: 0.7258
Epoch 8: train/loss: 3.7057 valid/loss: 2.4827 train/time: 12.86s valid/time: 2.78s train/acc: 0.4571 valid/acc: 0.6613
Early stopping!
Best model (epoch=3, step=21) saved to
RelPron/logs/best_model.lt
Training completed!
train/time: 1m45s train/time_per_epoch: 13.14s train/time_per_step: 1.88s valid/time: 23.12s valid/time_per_eval: 2.89s
Results¶
Finally, we visualise the results and evaluate the model on the test data.
import matplotlib.pyplot as plt
fig, ((ax_tl, ax_tr), (ax_bl, ax_br)) = plt.subplots(2, 2, sharex=True, sharey='row', figsize=(10, 6))
ax_tl.set_title('Training set')
ax_tr.set_title('Development set')
ax_bl.set_xlabel('Epochs')
ax_br.set_xlabel('Epochs')
ax_bl.set_ylabel('Accuracy')
ax_tl.set_ylabel('Loss')
colours = iter(plt.rcParams['axes.prop_cycle'].by_key()['color'])
range_ = np.arange(1, len(trainer.train_epoch_costs)+1)
ax_tl.plot(range_, trainer.train_epoch_costs, color=next(colours))
ax_bl.plot(range_, trainer.train_eval_results['acc'], color=next(colours))
ax_tr.plot(range_, trainer.val_costs, color=next(colours))
ax_br.plot(range_, trainer.val_eval_results['acc'], color=next(colours))
# mark best model as circle
best_epoch = np.argmax(trainer.val_eval_results['acc'])
ax_tl.plot(best_epoch + 1, trainer.train_epoch_costs[best_epoch], 'o', color='black', fillstyle='none')
ax_tr.plot(best_epoch + 1, trainer.val_costs[best_epoch], 'o', color='black', fillstyle='none')
ax_bl.plot(best_epoch + 1, trainer.train_eval_results['acc'][best_epoch], 'o', color='black', fillstyle='none')
ax_br.plot(best_epoch + 1, trainer.val_eval_results['acc'][best_epoch], 'o', color='black', fillstyle='none')
ax_br.text(best_epoch + 1.4, trainer.val_eval_results['acc'][best_epoch], 'early stopping', va='center')
# print test accuracy
model.load(trainer.log_dir + '/best_model.lt')
val_acc = acc(model(val_circuits), val_labels)
print('Validation accuracy:', val_acc.item())
Validation accuracy: 0.7419354838709677
See also: